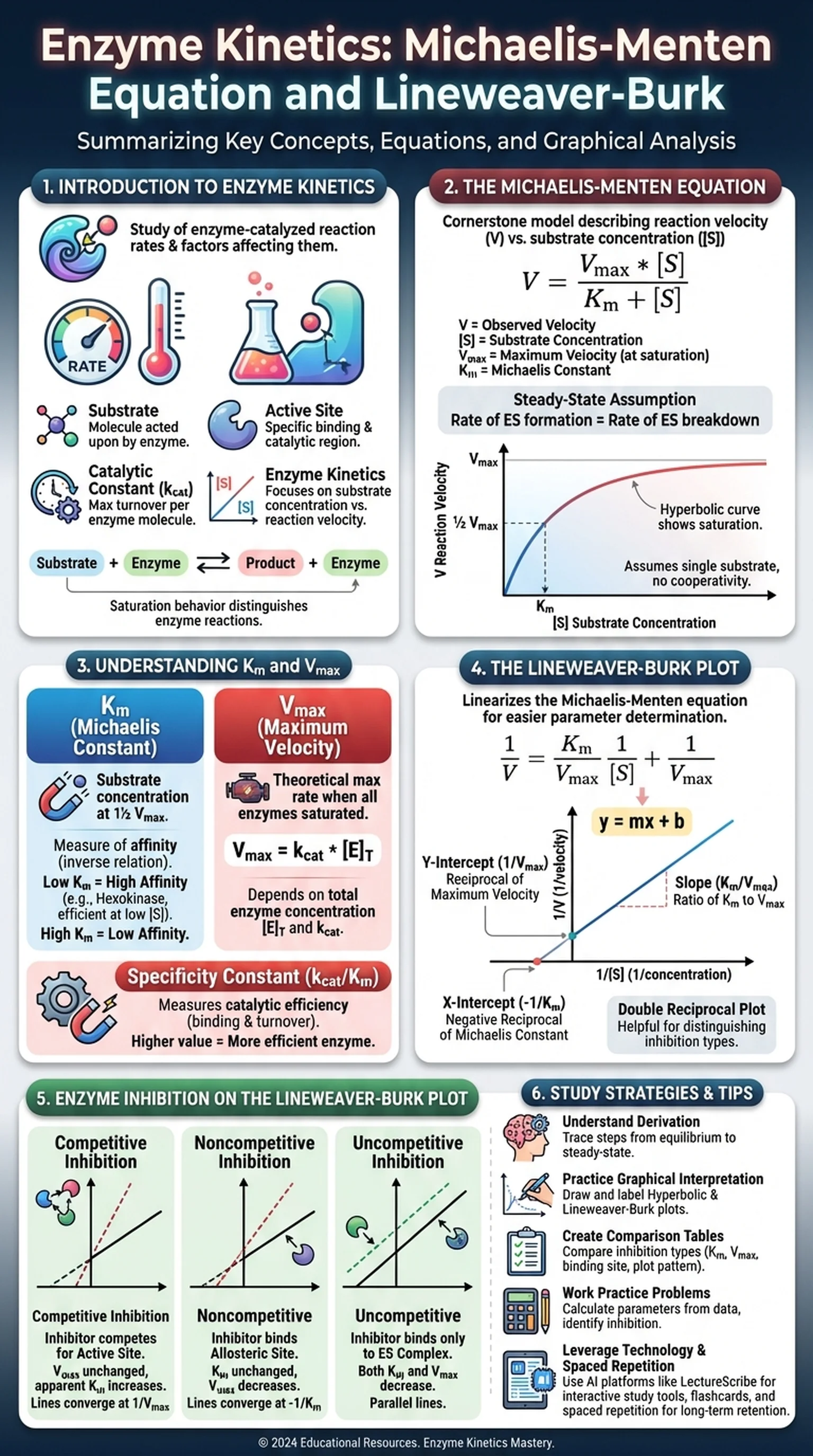
Introduction to Enzyme Kinetics
Enzyme kinetics is the branch of biochemistry that studies how fast enzyme-catalyzed reactions proceed and what factors influence their rates. By analyzing the speed at which substrates are converted into products, researchers can uncover the mechanisms by which enzymes operate, predict cellular behavior under different conditions, and design drugs that target specific enzymes. The quantitative framework of enzyme kinetics provides the foundation for understanding everything from metabolic flux in a living cell to the pharmacokinetics of therapeutic agents.
At its core, enzyme kinetics examines the relationship between substrate concentration and reaction velocity. When substrate is scarce, adding more substrate increases the reaction rate almost linearly. As substrate concentration rises, however, the rate of increase slows until the enzyme becomes saturated and the reaction reaches a maximum velocity. This saturation behavior distinguishes enzyme-catalyzed reactions from simple chemical reactions and reflects the finite number of enzyme active sites available to bind substrate at any given moment.
The study of enzyme kinetics also reveals how enzymes achieve their remarkable catalytic power. Enzymes lower the activation energy of reactions by stabilizing the transition state, and kinetic measurements allow scientists to quantify this effect. Parameters such as the catalytic constant (kcat), the Michaelis constant (Km), and the maximum velocity (Vmax) are all derived from kinetic experiments. Together, these values describe an enzyme's efficiency, its affinity for substrate, and the upper limit of its catalytic output. Understanding these parameters is essential for students preparing for the MCAT, USMLE, or any advanced biochemistry coursework.
Key Terms
The study of the rates of enzyme-catalyzed reactions and the factors that affect them, including substrate concentration, pH, and temperature.
The molecule upon which an enzyme acts, binding to the active site and being converted into product.
The specific region on an enzyme where the substrate binds and the catalytic reaction occurs.
The turnover number representing the maximum number of substrate molecules converted to product per enzyme molecule per unit time.