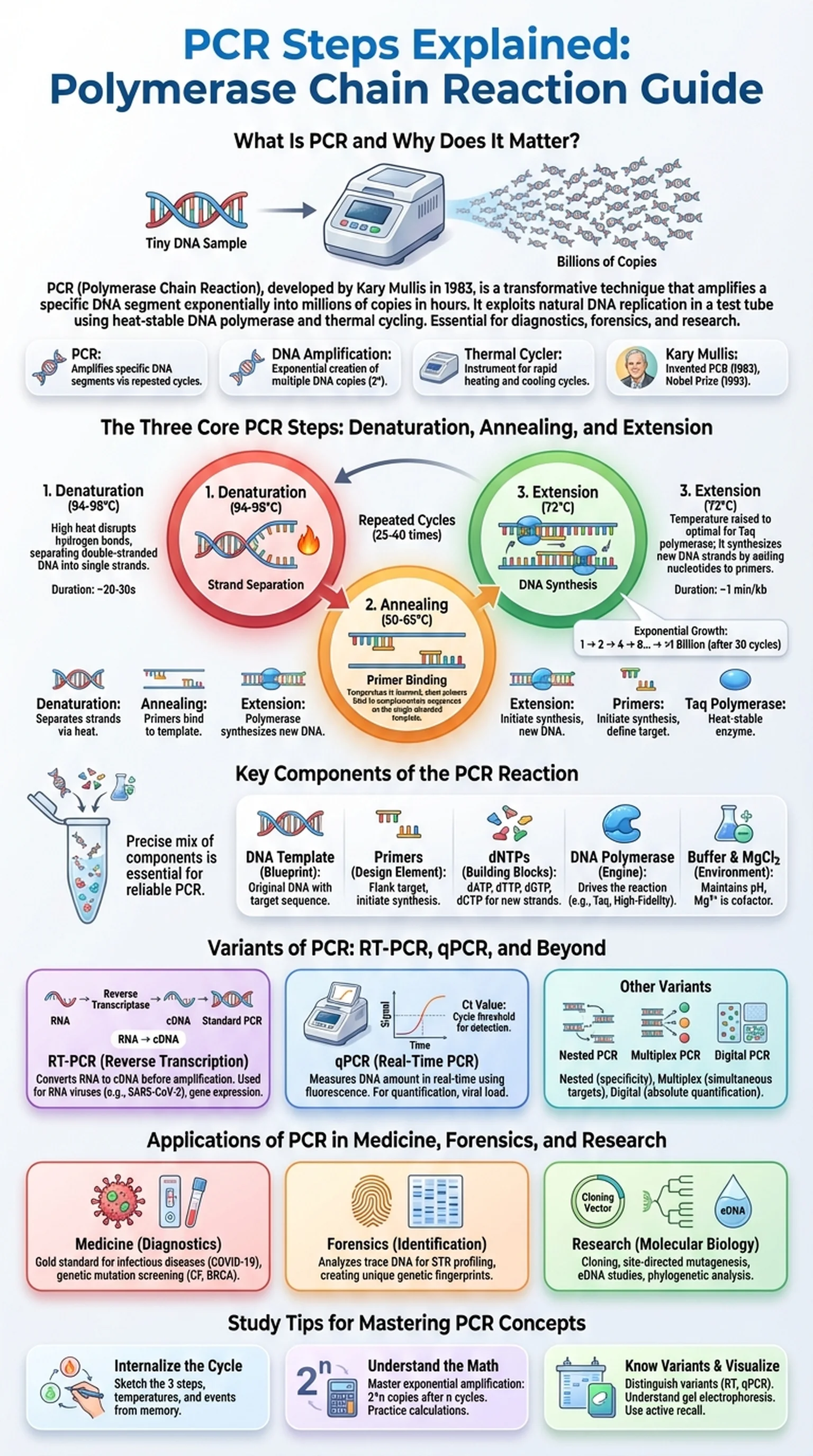
What Is PCR and Why Does It Matter?
PCR, or the polymerase chain reaction, is one of the most transformative techniques in modern molecular biology. Developed by Kary Mullis in 1983, PCR allows scientists to take a tiny sample of DNA and amplify it into millions or even billions of copies within just a few hours. This DNA amplification capability revolutionized fields ranging from forensic science and medical diagnostics to evolutionary biology and genetic engineering.
At its core, the PCR technique exploits the natural mechanism of DNA replication. In a living cell, enzymes unwind the double helix and synthesize new complementary strands. PCR mimics this process in a test tube by using a heat-stable DNA polymerase, short DNA primers, free nucleotides, and a thermal cycler that rapidly changes temperature. The elegance of the polymerase chain reaction lies in its exponential nature: each cycle doubles the number of target DNA molecules, so after 30 cycles a single molecule can theoretically become over one billion copies.
The importance of PCR cannot be overstated. Before its invention, obtaining enough DNA for analysis required laborious cloning procedures that took weeks. Today, the PCR technique is used in COVID-19 testing, paternity analysis, cancer mutation screening, and species identification from environmental samples. Understanding the PCR steps is essential for any student of biology, biochemistry, or medical sciences, as this foundational method appears on exams like the MCAT, GRE Biology, and AP Biology with remarkable frequency.
Key Terms
Polymerase chain reaction; a laboratory technique used to amplify specific segments of DNA through repeated cycles of denaturation, annealing, and extension.
The process of creating multiple copies of a specific DNA sequence, achieved exponentially through the polymerase chain reaction.
A laboratory instrument that rapidly heats and cools samples to the precise temperatures required for each step of the PCR technique.
The American biochemist who invented PCR in 1983 and was awarded the Nobel Prize in Chemistry in 1993 for this work.